Have you ever realized that at the very beginning, you started out as just one tiny cell. From that single cell, all the different parts of your body, like your brain cells, heart muscles, blood cells, even skin developed. The question is, how does that one original cell somehow “know” what to turn into? How does it decide to become a brain cell or a heart cell, instead of just staying the same?
It’s like you plant a single seed in the ground. Depending on where the seed ends up and the nutrients it receives, it can grow into a tree, a flower, or a vegetable. The same seed has the potential to become different plants, just like a single cell can become different types of cells in your body.
For decades, we’ve known that this process, cell fate determination, is influenced by two main things.
- First, there are clear biological rules guiding their development, like instructions you follow.
- Second, there’s randomness that happens by chance, which can change the outcome.
It can be compared to a game of Plinko, where a chip falls down a board. The drift is like gravity pulling the chip down a planned route, while the diffusion is like the chip bouncing randomly off pegs, sometimes ending up in unexpected places. Overall, the process involves both predictable guidance and randomness working together.
For instance, you are watering a plant gift that has multiple flowering buds. The plant has a natural instruction to focus on blooming the largest flower first (drift), but sometimes random factors like slight shifts in sunlight or tiny movements can cause the plant to open a different flower initially instead (diffusion).
Until recently, our best mathematical models were like a weather forecast that could tell you it was raining but couldn’t account for the wind. But a team from the Broad Institute of MIT and Harvard, Massachusetts General Hospital, and Harvard Medical School has just released a new AI framework that finally learns to “listen” to that biological noise.
The system is called scDiffEq, and it’s changing the way we look at how life builds itself.
The Data Explosion That Forced Biologists to Rethink Their Tools
To understand why this is a big deal, we have to look at how we’ve been studying cells lately.
We use a tool called single-cell RNA sequencing (scRNA-seq), this technique lets them see which genes are active in each individual cell, measuring thousands of genes at once. It’s a really useful method, but it creates a huge amount of data to analyze.
Traditional math models (differential equations) have been the backbone of physics for a century, but they hit a wall when trying to describe the complex, high-dimensional reality of 20,000 genes interacting at once.
“Neural differential equations changed this”, explains Michael Vinyard, the lead author of the study published in Nature Machine Intelligence. By using neural networks to “learn” the equations directly from the data, researchers can finally map the complex “vector field” of a cell’s life.
scDiffEq Treats Biological Noise Differently
Traditional machine learning models assume that randomness or “noise” in biological data stays the same everywhere, like static on a radio that’s always at the same volume. But scDiffEq doesn’t make that assumption. Instead, it understands that different cell states can have different amounts of randomness, meaning the noise level can change depending on what kind of cell you’re looking at.
Key Breakthroughs of the Framework:
- Variable Stochasticity: A “progenitor” cell (a sort of biological “blank slate”) might be much noisier and more erratic than a cell that has already decided to become a blood cell.
- Drift vs. Diffusion: The model uses neural networks to map both the deterministic “drift” (the internal program) and the stochastic “diffusion” (the random fluctuations).
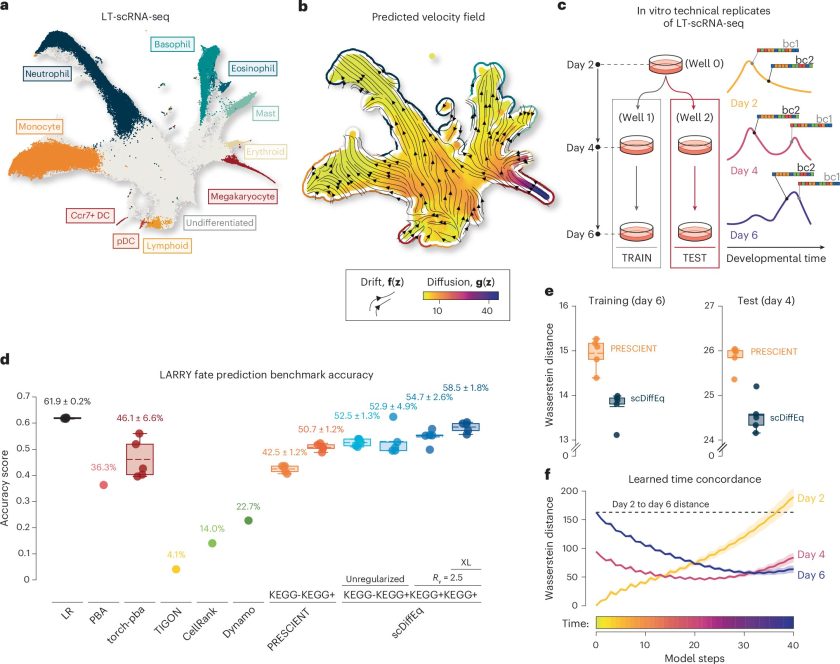
- Better Accuracy: When tested on blood cell development, scDiffEq predicted cell fates with 58% accuracy, an 8% jump over previous models.
This AI Model Is More Than Just a Prediction Tool
This research is exciting not only because it can predict what a cell might do next, but also because it lets scientists test different situations on a computer. They have a mathematical model that shows how cells develop and change.
Using this, they can simulate what would happen if they changed a gene or added a drug, without doing real experiments. It’s like running virtual tests to see how cells would respond to different conditions before trying them in real life.
“Deep computational modeling… could be used to predict the effect of perturbations, which is critical for finding drug targets and designing clinical interventions”, says Gad Getz, co-senior author of the paper.
This has massive implications for diseases like cancer. Many cancers are essentially “hijacked” developmental processes where mutations break the decision-making circuits of the cell. If we can model exactly where those circuits go wrong, we can find better ways to fix them.
Takeaway
The model reduces complex genetic information into a simpler form because analyzing all over 20,000 genes separately is very hard and requires a lot of computing power. In the future, they want to:
- Scale up to map genes directly and more reliably.
- Integrate more data layers, such as chromatin accessibility, to see not just which genes are active, but which ones are “open” for business.
- Incorporate CRISPR data (specifically from “Perturb-seq” experiments) to dramatically improve how the AI predicts the effects of genetic changes.
We often think of biology as a clockwork machine, but scDiffEq reminds us that life is actually a bit more like a storm, a mixture of predictable currents and random gusts. By building an AI that finally understands the “noisiness” of development, we aren’t just getting better at predicting where a cell is going. We’re getting closer to being able to steer it.
Whether it’s finding a new drug target for a rare disease or understanding how to guide stem cells to repair a damaged heart, the ability to model the “Plinko board” of our own biology is a giant leap forward.
Frequently Asked Questions (FAQs)
1. What is cell fate determination in biology?
Cell fate determination is the process by which a single cell decides what type of specialized cell it will become, such as a brain cell, blood cell, or muscle cell. This decision is influenced by genetic instructions and random biological fluctuations during development.
2. How does AI help scientists understand cell development?
AI helps scientists analyze massive single-cell datasets by learning complex patterns that traditional mathematical models cannot capture. New AI frameworks like scDiffEq can model both predictable genetic programs and random biological noise to better predict how cells change over time.
3. What is scDiffEq and why is it important?
scDiffEq is an AI-based neural differential equation framework developed by researchers at the Broad Institute, MIT, and Harvard. It is important because it accurately models both deterministic and stochastic forces in cell development, improving predictions of cell fate decisions.
4. What is biological “noise” and why does it matter?
Biological noise refers to random fluctuations in gene activity within cells. This noise matters because it can influence cell fate outcomes, especially in early or progenitor cells, and ignoring it can lead to inaccurate biological models.
5. How is scDiffEq different from traditional cell modeling methods?
Unlike traditional models that assume constant randomness, scDiffEq allows noise levels to vary across different cell states. This makes it more accurate for modeling real biological systems where uncertainty changes as cells specialize.
6. Can scDiffEq help with drug discovery and disease research?
Yes. scDiffEq enables in silico experiments that simulate gene changes or drug treatments, helping researchers predict how cells will respond. This is especially valuable for cancer research and precision medicine.
7. Why is single-cell RNA sequencing important for AI models like scDiffEq?
Single-cell RNA sequencing provides detailed gene activity data for individual cells, which AI models use to learn developmental trajectories. Without scRNA-seq data, frameworks like scDiffEq would not be able to map cell fate decisions accurately.
Source: Phys.org